Tutorial: Data Analysis¶
This is the follow up to Tutorial: Data Collection. We have measured bitstrings for the single-qubit circuit \(R_y(\theta)\) for various thetas. In this analysis, we compute \(\langle Z \rangle (\theta)\), compare to the anayltically expected true value, and fit to a depolarizing noise model with T1 decay during readout.
Loading data¶
We can use utilities in ReCirq to query the filesystem and load in a dataset. Please recall that all tasks have an associated EXPERIMENT_NAME and a dataset_id which define the top two hierarchies in the filesystem. We import these values from the data collection script to ensure consistency.
[1]:
import cirq
import recirq
from recirq.readout_scan.tasks import EXPERIMENT_NAME, DEFAULT_BASE_DIR
recirq.iterload_records uses these two bits of information to iterate over records saved using recirq.save (in the data collection script.
This also gives you a chance to do post-processing on the data. In general, you should do some massaging of the data and put the results into a pandas DataFrame. DataFrames are great for doing statistics and visualizations across tabular data.
[2]:
import numpy as np
import pandas as pd
records = []
# Load all data, do some light processing
for record in recirq.iterload_records(dataset_id='2020-02-tutorial', base_dir=DEFAULT_BASE_DIR):
# Expand task dataclass into columns
recirq.flatten_dataclass_into_record(record, 'task')
# Unwrap BitArray into np.ndarray
all_bitstrings = [ba.bits for ba in record['all_bitstrings']]
# Compute <Z>
record['z_vals'] = [np.mean((-1)**bitstrings, axis=0).item() for bitstrings in all_bitstrings]
# Don't need to carry around the full array of bits anymore
del record['all_bitstrings']
records.append(record)
df = pd.DataFrame(records)
print(len(df))
df.head()
5
[2]:
| timestamp | thetas | dataset_id | device_name | n_shots | qubit | resolution_factor | z_vals | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2020-08-06T21:57:59.931054 | [-1.5707963267948966, -1.3089969389957472, -1.... | 2020-02-tutorial | Syc23-simulator | 40000 | (3, 2) | 6 | [-0.00355, 0.2521, 0.4957, 0.71, 0.8579, 0.958... |
| 1 | 2020-08-06T21:58:12.502284 | [-1.5707963267948966, -1.3089969389957472, -1.... | 2020-02-tutorial | Syc23-simulator | 40000 | (4, 3) | 6 | [-0.00295, 0.2551, 0.49295, 0.70525, 0.86315, ... |
| 2 | 2020-08-06T21:58:16.949905 | [-1.5707963267948966, -1.3089969389957472, -1.... | 2020-02-tutorial | Syc23-simulator | 40000 | (5, 0) | 6 | [0.0044, 0.2568, 0.4977, 0.69205, 0.8605, 0.95... |
| 3 | 2020-08-06T21:58:08.485639 | [-1.5707963267948966, -1.3089969389957472, -1.... | 2020-02-tutorial | Syc23-simulator | 40000 | (4, 2) | 6 | [0.01045, 0.253, 0.49825, 0.70115, 0.8622, 0.9... |
| 4 | 2020-08-06T21:58:04.450974 | [-1.5707963267948966, -1.3089969389957472, -1.... | 2020-02-tutorial | Syc23-simulator | 40000 | (4, 1) | 6 | [-0.0006, 0.2591, 0.4936, 0.7041, 0.85785, 0.9... |
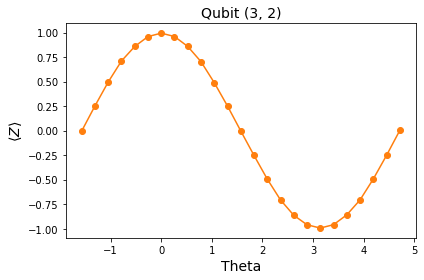
Plot the data¶
A good first step.
[3]:
%matplotlib inline
from matplotlib import pyplot as plt
entry = df.iloc[0] # Pick the first qubit
plt.plot([], []) # advance color cycle in anticipation of future analysis
plt.plot(entry['thetas'], entry['z_vals'], 'o-')
plt.xlabel('Theta', fontsize=14)
plt.ylabel(r'$\langle Z \rangle$', fontsize=14)
plt.title("Qubit {}".format(entry['qubit']), fontsize=14)
plt.tight_layout()
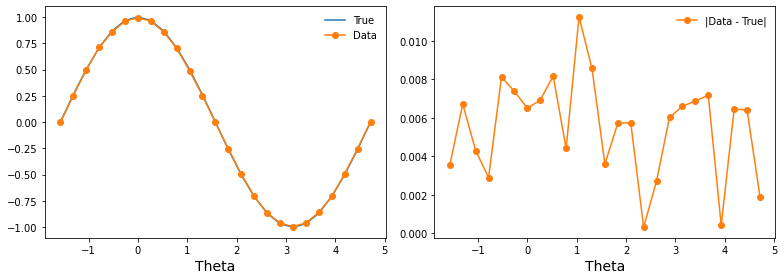
How does it compare to analytical results?¶
You could imagine setting up a separate task for computing and saving analytic results. For this single qubit example, we’ll just compute it on the fly.
[4]:
qubit = cirq.LineQubit(0)
thetas = df.iloc[0]['thetas']
class _DummyMeasurementGate(cirq.IdentityGate):
"""A dummy measurement used to trick simulators into applying
readout error when using PauliString.expectation_from_xxx."""
def _measurement_key_(self):
return 'dummy!'
def __repr__(self):
if self.num_qubits() == 1:
return '_DummyMeasurementGate'
return '_DummyMeasurementGate({!r})'.format(self.num_qubits())
def __str__(self):
if (self.num_qubits() == 1):
return 'dummyM'
else:
return 'dummyM({})'.format(self.num_qubits())
def _circuit_diagram_info_(self, args):
from cirq import protocols
return protocols.CircuitDiagramInfo(
wire_symbols=('dM',) * self.num_qubits(), connected=True)
def dummy_measure(qubits):
return _DummyMeasurementGate(num_qubits=len(qubits)).on(*qubits)
def get_circuit(theta):
return cirq.Circuit([
cirq.ry(theta).on(qubit),
dummy_measure([qubit])
])
true_z_vals = []
for theta in thetas:
wf = cirq.final_wavefunction(get_circuit(theta))
op = cirq.Z(qubit) * 1.
true_z_val = op.expectation_from_wavefunction(wf, qubit_map={qubit:0}, check_preconditions=False)
true_z_vals.append(np.real_if_close(true_z_val).item())
true_z_vals = np.array(true_z_vals)
true_z_vals
[4]:
array([-1.26880515e-08, 2.58819014e-01, 5.00000000e-01, 7.07106769e-01,
8.66025388e-01, 9.65925872e-01, 1.00000000e+00, 9.65925872e-01,
8.66025388e-01, 7.07106769e-01, 5.00000000e-01, 2.58819014e-01,
-1.26880515e-08, -2.58819014e-01, -4.99999970e-01, -7.07106709e-01,
-8.66025388e-01, -9.65925872e-01, -1.00000000e+00, -9.65925872e-01,
-8.66025388e-01, -7.07106709e-01, -4.99999970e-01, -2.58819014e-01,
-1.26880515e-08])
[5]:
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(11, 4))
ax1.plot(thetas, true_z_vals, '-', label='True')
ax1.plot(entry['thetas'], entry['z_vals'], 'o-', label='Data')
ax2.plot([], []) # advance color cycle
ax2.plot(entry['thetas'], np.abs(true_z_vals - entry['z_vals']), 'o-', label='|Data - True|')
ax1.legend(loc='best', frameon=False)
ax2.legend(loc='best', frameon=False)
ax1.set_xlabel('Theta', fontsize=14)
ax2.set_xlabel('Theta', fontsize=14)
fig.tight_layout()
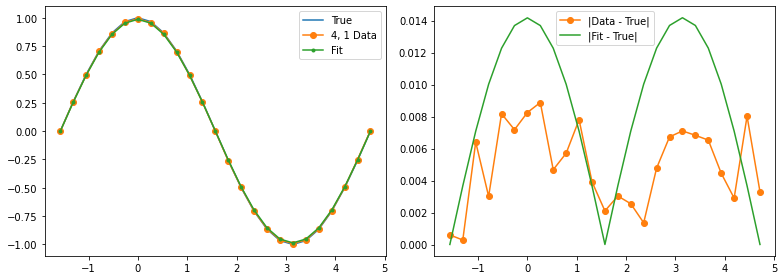
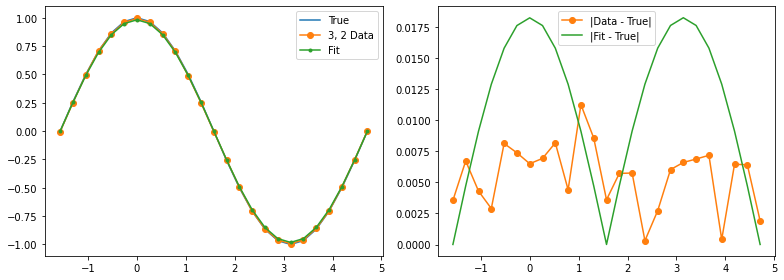
Learn a model¶
Our experimental data has some wiggles in it, but it also has a clear pattern of deviation from the true values. We can hypothesize a (parameterized) noise model and then use function minimization to fit the noise model parameters.
[6]:
import scipy.optimize
import cirq.contrib.noise_models as ccn
def get_obj_func(data_expectations):
all_results = []
def obj_func(x):
depol_prob, decay_prob, readout_prob = x
if depol_prob < 0 or decay_prob < 0 or readout_prob < 0:
# emulate constraints by returning a high cost if we
# stray into invalid territory
return 1000
sim = cirq.DensityMatrixSimulator(
noise=ccn.DepolarizingWithDampedReadoutNoiseModel(
depol_prob=depol_prob, decay_prob=decay_prob, bitflip_prob=readout_prob))
results = []
for theta in thetas:
density_result = sim.simulate(get_circuit(theta))
op = cirq.Z(qubit) * 1.
true_z_val = op.expectation_from_wavefunction(density_result.final_density_matrix, qubit_map=density_result.qubit_map, check_preconditions=False)
results.append(np.real_if_close(true_z_val).item())
results = np.array(results)
all_results.append(results)
cost = np.sum(np.abs(results - data_expectations))
return cost
return obj_func, all_results
[7]:
def print_result(x):
depol_prob, decay_prob, readout_prob = x
print(f'depol = {depol_prob:.2%}')
print(f'decay = {decay_prob:.2%}')
print(f'readout = {readout_prob:.2%}')
[8]:
dfb = df
dfb = dfb.head(5) # Remove this to do all qubits
len(dfb)
[8]:
5
[9]:
# Initial values
depol_prob = 0.01
decay_prob = 0.01
readout_prob = 0.01
opt_results = []
for i, entry in dfb.iterrows():
ofunc, results = get_obj_func(entry['z_vals'])
opt_result = scipy.optimize.minimize(ofunc,
[depol_prob, decay_prob, readout_prob],
method='nelder-mead',
options={'disp': True})
label = f"{entry['qubit'].row}, {entry['qubit'].col}"
print("Qubit", label)
print_result(opt_result.x)
opt_results.append(opt_result)
data_expectations = entry['z_vals']
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(11, 4))
ax1.plot(thetas, true_z_vals, label='True')
ax1.plot(thetas, data_expectations, 'o-', label=f'{label} Data')
ax1.plot(thetas, results[-1], '.-', label='Fit')
ax2.plot([], []) # advance color cycle
ax2.plot(thetas, np.abs(true_z_vals - data_expectations), 'o-', label='|Data - True|')
ax2.plot(thetas, np.abs(true_z_vals - results[-1]), '-', label='|Fit - True|')
ax1.legend(loc='best')
ax2.legend(loc='best')
fig.tight_layout()
plt.show()
Optimization terminated successfully.
Current function value: 0.183489
Iterations: 70
Function evaluations: 133
Qubit 3, 2
depol = 1.37%
decay = 0.00%
readout = 0.00%
Optimization terminated successfully.
Current function value: 0.206460
Iterations: 67
Function evaluations: 128
Qubit 4, 3
depol = 1.37%
decay = 0.00%
readout = 0.00%
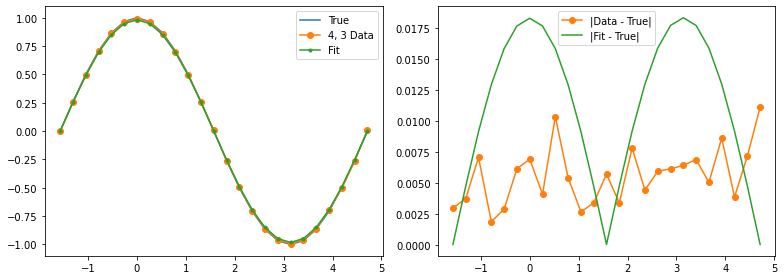
Optimization terminated successfully.
Current function value: 0.175666
Iterations: 69
Function evaluations: 135
Qubit 5, 0
depol = 1.37%
decay = 0.00%
readout = 0.00%
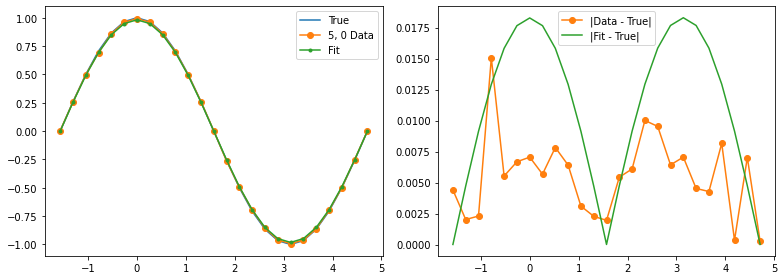
Optimization terminated successfully.
Current function value: 0.189504
Iterations: 61
Function evaluations: 119
Qubit 4, 2
depol = 1.31%
decay = 0.00%
readout = 0.06%
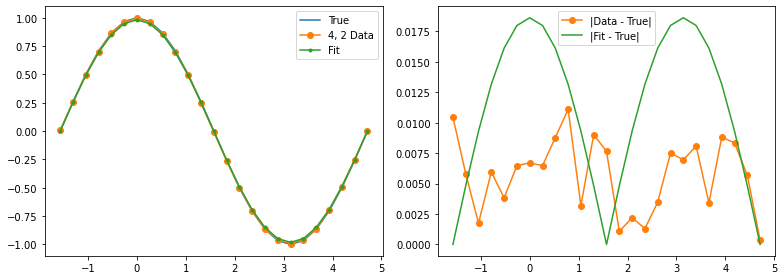
Optimization terminated successfully.
Current function value: 0.119917
Iterations: 87
Function evaluations: 167
Qubit 4, 1
depol = 1.06%
decay = 0.00%
readout = 0.00%